To date, 102 candidate COVID-19 vaccines are in clinical development and 184 in preclinical development, by use of a range of vaccine platforms (WHO). An efficacious COVID-19 vaccine could reduce the likelihood of infection, severity of disease, or degree of transmission within a population. It is imperative that accurate assessment of vaccine performance is carried out, especially to evaluate the long-term protection.
Currently, anti-N and anti-S antibody assays are used with the deliberate intention of identifying natural infection as opposed to the effects of vaccine. However, both assays have limitations. It was observed that significant number of cases (5%) react only to the spike or nucleocapsid antigens, but not both. Also, accumulating mutations driven by antigenic drift, or by selection will further reduce sensitivity of the existing tests. Therefore, traditional antibody assays using either the spike or nucleocapsid as antigens are limited in their detection accuracy.
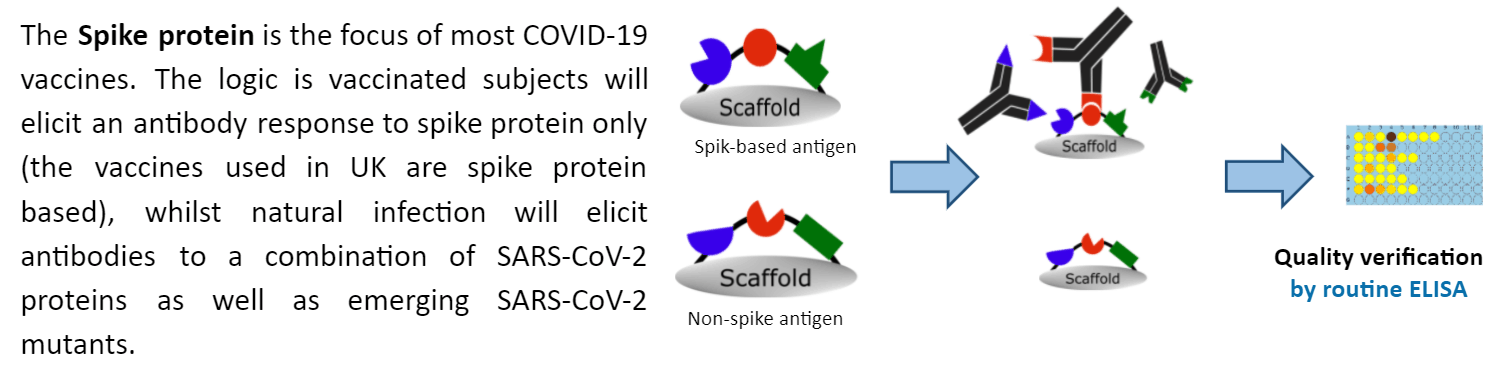
The Differential EpitoGen® COVID-19 test offers the ability to differentiate between humoral responses to vaccination and primary infection with SARS-CoV-2.
Principles of Differential EpitoGen® COVID-19 Test
- Appreciation of the B-cell epitope immunodominance phenomenon. This will enrich for the positive signal and improve sensitivity. Another advantage is that cross-reactive epitopes, are eliminated, subsequently improving specificity.
- Differential EpitoGen overcomes population heterogeneity (i.e. HLA genetic variation, co-morbidities, age, ethnicity, etc) by selecting those epitopes prevalent in the population.
- Differential EpitoGen amenable to combining multiple SARS-CoV-2 proteins in one test, subsequently improving sensitivity of the assay.
- Differential EpitoGen overcomes SARS-CoV-2 genetic variability (i.e. emerging mutations) by inclusion of prevalent mutations.
Intended Use of EpitoGen® Differential
-
Differentiate between vaccine-induced and infection-induced immunity
-
Offers a readout of Spike antibodies post vaccination/infection to evaluate immune memory
How EpitoGen® Differential was developed?
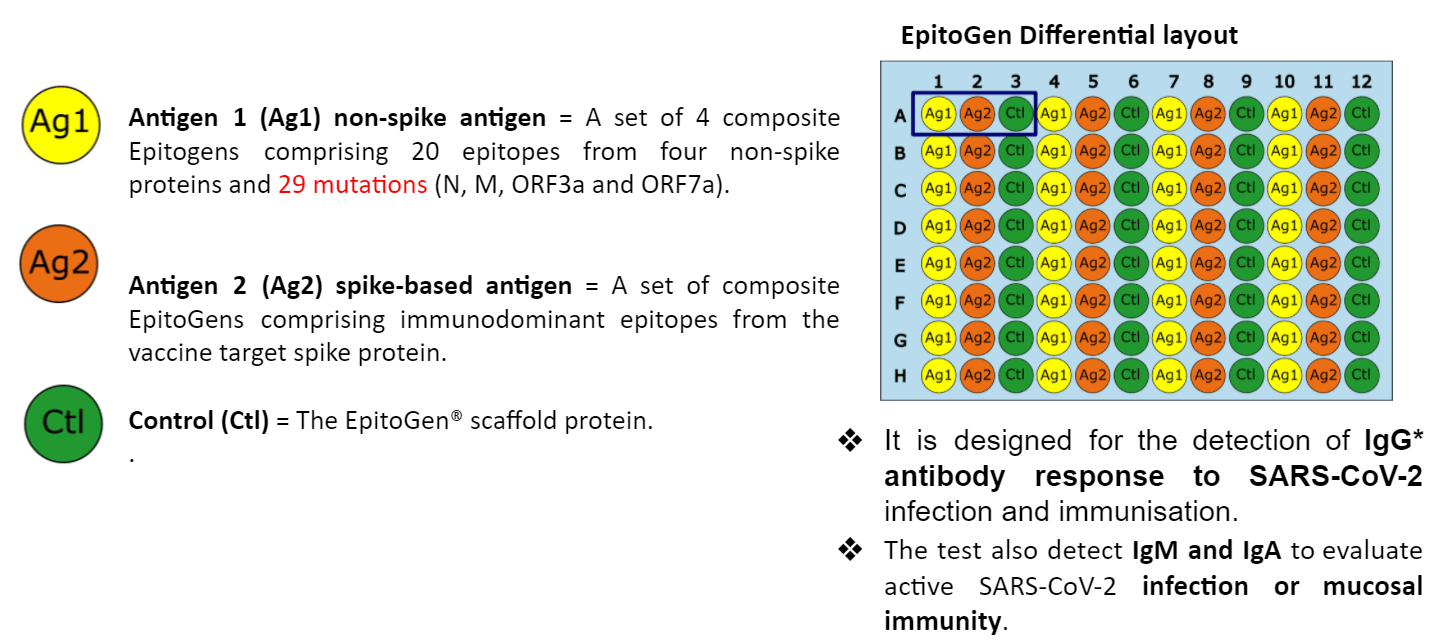
Using EpitoGen Technology, a set of 10 immunodominant non-spike reference epitopes and 10 non-spike mutant epitopes corresponding to SARS-CoV-2 viral proteins were complexed to create a non-spike antigen. Whilst a set of reference spike immunodominant epitopes were complexed to create spike-based antigen.
For purchase orders, please contact us at info@vertebrateantibodies.com
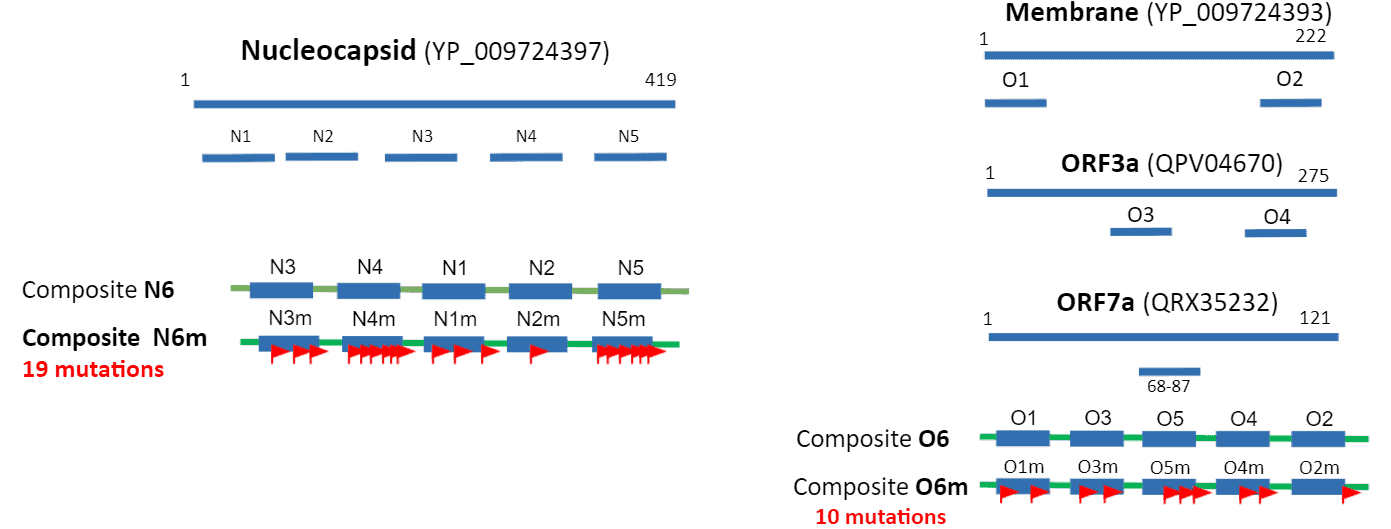
- Immunogenicity profiling of SARS-CoV-2 identified N, M, ORF3a, ORF7a as most antigenic viral proteins.
- Immunodominant epitopes were identified using EpitopePredikt, then selected epitopes fused together and expressed on EpitoGen® scaffold.
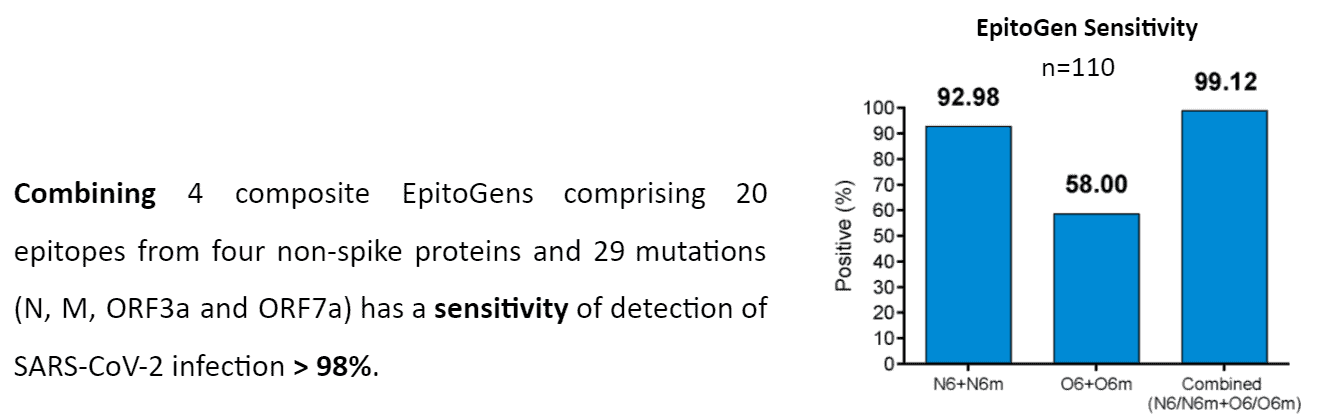
- Prevalent mutants were included to enhance the detection accuracy of the non-spike complex. Some mutations will create new epitopes to which specific antibody response is elicited. We included 29 prevalent mutations into the EpitoGen® Differential to enhance sensitivity.
-Nucleocapsid (N6m) mutations: S33I, A35V, P46S, D103Y, A152S, A156S, P168S, G125V, Q229H, M234I, S235F, T247I, A251V, T366I, K373N, D377Y, P383S, T391I, D401Y.
–Membrane mutations: D3G, K15R, D209Y.
–ORF3a mutations: K136E, A143S, Q213K, T223I
-ORF7a mutations: H73Y, R80I, P84S